The National Centre for Text Mining: A Vision for the Future
One of the defining challenges of e-Science is dealing with the data deluge [1] information overload and information overlook. More than 8,000 scientific papers are published every week (on Google Scholar, for example). Without sophisticated new tools, researchers will be unable to keep abreast of developments in their field and valuable new sources of research data will be under-exploited. The capability of text mining (TM) to find knowledge hidden in text and to present it in a concise form makes it an essential part of any strategy for addressing these problems. As text mining matures, it will increasingly enable researchers to collect, maintain, interpret, curate, and discover knowledge needed for research and education, efficiently and systematically. The National Centre for Text Mining (NaCTeM) is playing a critical role in ensuring that UK researchers are aware of and have access to effective TM solutions, and are able to exploit their capabilities to the full.
Text Mining Services for the UK Academic Community
There are many kinds of text mining services, with many sub-processes involved, and many types of user in varying domains with different needs, interests, purposes, requirements and degrees of technological aptitude.
NaCTeM has developed a number of high-quality text mining service exemplars for the UK academic community. The exemplars produced are:
TerMine, AcroMine [2][3] extract candidate terms from text, and map them between biology acronyms and their full forms. TerMine is a foundational service in that terminology is central to many activities and is often a hurdle for humans and language processors alike. Termine is a service for automatic term recognition which identifies the most important terms in a document ranking them according to their significance and proposing potential expansions of all acronyms found. It is based on the C-value method for automatic term recognition [4].
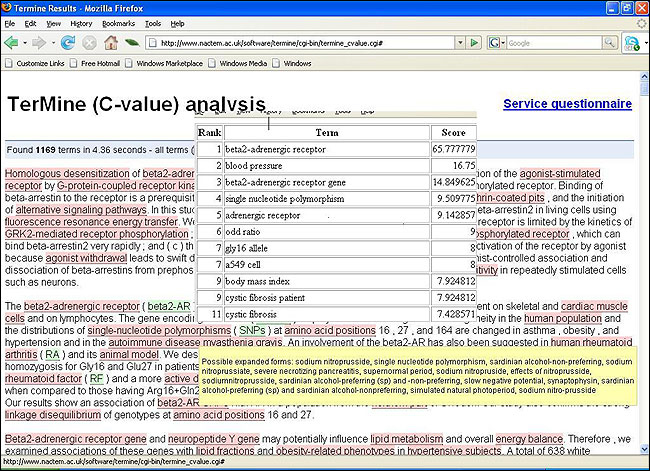
Figure 1: TerMine: A service for automatic term recognition
AcroMine is based on a novel approach for recognising acronym definitions in a text collection [5]. Applied to the whole MEDLINE, it deals with terminological variation, an integral part of the linguistic ability to realise a concept in different ways. This is also an obstacle to information retrieval.
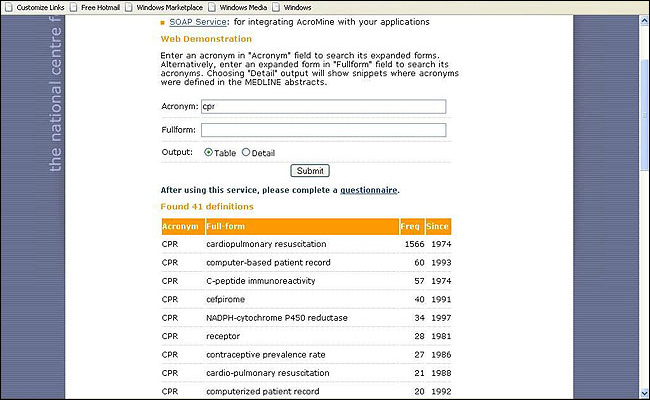
Figure 2: AcroMine finds expanded forms of acronyms and disambiguates them
AcroMine finds expanded forms of acronyms from a database created from the whole of Medline and disambiguates them. AcroMine has also been incorporated into the TerMine user interface. TerMine, moreover, crucially supports Semantic Web activities and has been used as an aid to ontology construction and controlled vocabularies [6].
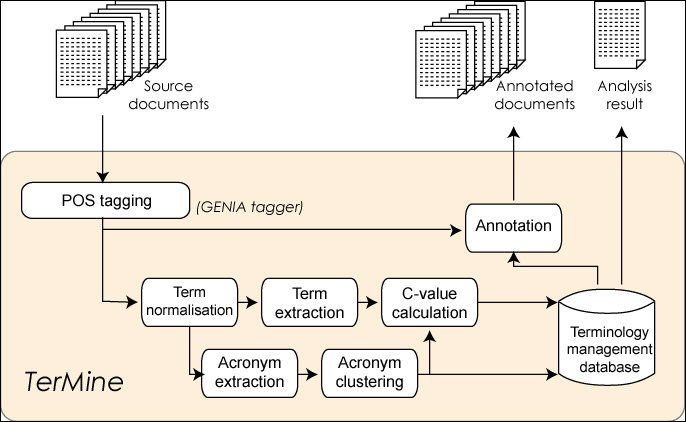
Figure 3: Overall architecture of TerMine
Cheshire/TerMine [7] integrates information retrieval (provided by Cheshire) [8] and TerMine to offer users a search facility based on terms with which they are familiar, and which moreover retrieves documents according to the importance of the terminology they contain. For example, this service allows the user to search for documents related to a subject area, to find associated terms related with the query and to select the most appropriate documents to view. This service identifies important terms (that the user may not have known) combining the best of search and browse models of information access.
MEDIE [9] provides real-time semantic information retrieval based on the retrieval of relational concepts from huge texts. It is an intelligent search engine which uses semantic retrieval technologies to identify sentences containing biomedical correlations for queried terms from Medline abstracts. The service runs on the whole of Medline and is based on semantically annotated texts using deep parsing and named entity recognition. Sentences are annotated in advance with semantic structures and stored in a structured database. User requests are converted on the fly into patterns of these semantic annotations, and texts are retrieved by matching these patterns with the pre-computed semantic annotations [10].
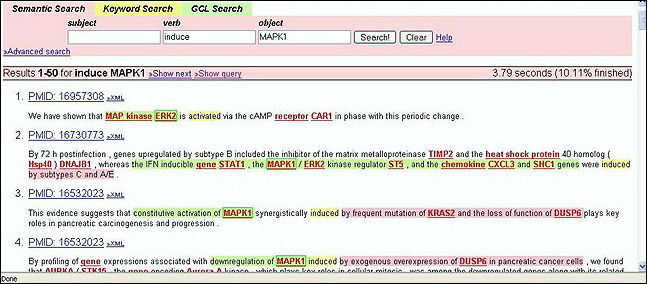
Figure 4: Medie: An intelligent search engine using semantic retrieval technologies
InfoPubMed [11] extracts and visualises protein-protein interactions. Info-PubMed is an efficient PubMed search tool, helping users to find information about biomedical entities such as genes, proteins, and the interactions between them.
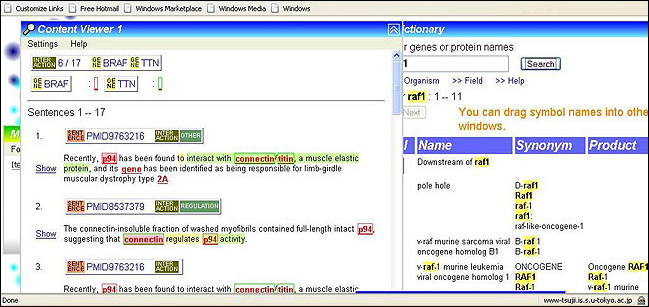
Figure 5: InfoPubMed provides information and visualisation of biomedical interactions extracted from Medline
It provides information and visualisation of biomedical interactions extracted from Medline using deep semantic parsing technology. This is supplemented with the GENA term dictionary which consists of over 200,000 protein/gene names and identifications of disease types and organisms
NaCTeM services are underpinned by a number of generic natural language processing tools which have already been adapted for the domain of biology. Most of these tools are currently being adapted for use in the social sciences within the framework of the ASSERT [12][13 Project whose aim is to provide automated assistance to social scientists undertaking systematic reviews of the research literature. Figure 6 shows how text mining techniques are being used to support the stages of searching, screening and synthesising in systematic reviews.
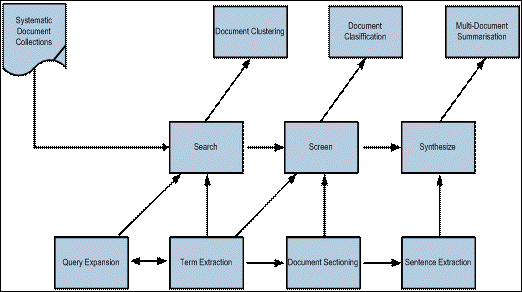
Figure 6: Text mining technologies for enhancing systematic reviewing
Text mining techniques have the potential to revolutionise the way we approach research synthesis but our longer term interest is to understand how we can apply these techniques more widely in a variety of use cases in the social sciences. To achieve this, we will use systematic reviewing to demonstrate the potential of text mining for the social science research community and to establish requirements for a generic toolkit of text mining services which can be integrated into different research practices. This provides its own set of issues for development in terms of interoperability with techniques or software currently employed in the systematic review activity but also with other text mining tools and services used by the social science community. For example a researcher investigating the role of new media in politics could be interested in combining the toolset with Internet news feed or blog readers, their own evidence tracking systems or even other tools for carrying out opinion analysis. We need to ensure that our tools are therefore flexible and robust enough to allow for this, whilst providing sufficient functionality to ensure interoperability between the many formats and standards that this would entail.
The NaCTeM Roadmap
Based on the current service exemplars described previously, NaCTeM's roadmap in figure 7 below outlines our vision for the next 5 years. This vision situates NaCTeM with respect to:
- the general goals text mining aims to address
- the main scientific challenges it helps to solve, thus providing breakthrough to scientific discoveries across domains, and
- the main issues related to the deployment, use and uptake of NaCTeM text mining solutions and services in respect of a wide variety of users. The roadmap sets out the core technology underpinning the provision of NaCTeM's text mining services and solutions over this period. As our services are user-driven these examples will be further refined and developed in close consultation with our user communities.
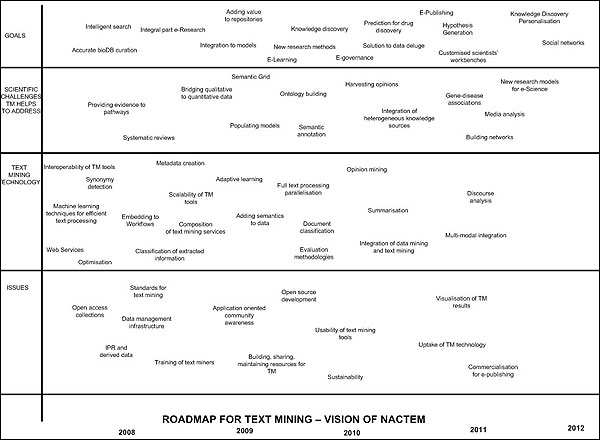
Figure 7: The NaCTeM Roadmap (larger version)
Issues
Full paper processing is necessary for the discovery of new knowledge and evidence from literature, but currently there is limited availability of open access collections for the scientific community and there is uncertainty over IPR (Intellectual Property Rights) in data derived via text mining [14], especially as publishers move to new models [15][16]. Standards are needed for the types of annotation we use to represent layers of linguistic and semantic analysis, and thus there is a need for cooperation among stakeholders. TM tools are increasingly released as open source, but in order to be interoperable, common infrastructure and annotation schemes [17] have to be adopted. Large-scale resources (annotated corpora, lexicons, ontologies) [18] are required to support TM, but are expensive for one centre to produce (and maintain), requiring collaboration, sharing and co-funding. TM as a new technology faces potential barriers which must be tackled. TM tools must match users' requirements and be usable if the technology is to achieve wide adoption. NaCTeM is working closely with NCeSS [19] to ensure that it is in a position to identify barriers to adoption and develop strategies to address them.
Text Mining Technology Supporting Service Provision
An important aspect of the roadmap is the core technology and computational infrastructure needed to allow NaCTeM text-mining services to be developed for use by the community. Our roadmap lays out the main steps we expect to take. These steps are, in several cases, related to general rather than specific technological development. For example, more generally applicable text mining technology to support large-scale services depends on other developments, e.g. the ability to process full texts [20]. In order to process full texts on a large scale to support user services, a pre-requisite is parallelisation. Early steps in being able to deploy our text mining services as Web Services lead to processes involving embedding text mining Web Services in workflows, composition of TM services via Web Services, and so on.
Development of highly efficient machine-learning techniques for text processing is a pre-requisite for classification of extracted information that forms the core of large-scale (repository-wide) metadata creation. The ability more generally to add semantics to textual data on a large scale supports full-text processing. In order to be able to adapt rapidly to new domains in coming years, adaptive learning technology must be in place. Integration of data mining and text mining on a large scale becomes possible at a later stage as our enabling tools and services are put in place.
None of the above stages can happen easily without early effort to provide workable levels of interoperability, re-usability and portability. While NaCTeM will continue to offer a portal enabling users to find out about best-of-breed text mining tools, integration of arbitrary tools demands not only a common infrastructure but also a common linguistic annotation scheme [21]. Although there are solutions for the former there is no solution for the latter. NaCTeM has actively promoted the idea of interoperability of text mining tools in the wider community. We are already using IBM's UIMA (Unstructured Information Management Architecture) [22] to support interoperability. UIMA has a set of useful functionalities, such as type definitions shared by modules, management of complex objects, links between multiple annotations and the original text, and a GUI for module integration. However, it simply provides a framework for, and not the detail of, common annotation schemes. Thus, the user community has to develop its own platforms with a set of actual software modules. In addition, simply wrapping existing modules in UIMA does not offer a complete solution for flexible tool integration, necessary for practical applications in a variety of domains. Users, and this includes both developers and end-users of TM systems, tend to be confused when faced with choosing appropriate modules for their own tasks from a collection of a large number of tools.
NaCTeM has ensured that its own tools and services are interoperable thus providing users with a coherent interoperable text mining suite. In addition, we have developed a general combinatorial text mining comparator which: generates possible combinations of tools for a specific workflow; compares/evaluates the results; facilitates module integration; and guides the selection of text mining modules [23] Processing full texts on a large scale necessitates the development of an appropriate massively parallel data management infrastructure to support the use of the machine learning text mining tools we use to improve the accuracy of our services and the ease of adaptation of our toolkit for new domains. Serious use of text mining services implies the ability and the capacity of such services to handle very large amounts of data if e-scientists are to make the kinds of breakthrough they are hoping for. It is part of the remit of NaCTeM to support such large scale processing. Taken together with the issue of interoperability this further motivates us to provide services around the tools we develop. The integration of TM into a workflow environment (e.g. Taverna, [24]) will allow us to reach a wide community of e-scientists and will provide additional functionality to the widely popular workflow environment by augmenting data with evidence and results from the literature.
Scientific Challenges NaCTem Services Help to Address
The challenge of providing evidence for pathways from literature is currently being tackled within the REFINE Project [25]. Our aim is to annotate texts semantically in order to extract biological relations which provide the evidence for the detection and classification of the species and reactions in bio-chemical models.
Although we cannot provide the full answer to the challenge of extracting quantitative data from qualitative materials which is currently exercising the social sciences, we are currently offering ways of eliminating some of its routine and mechanical aspects [12]. Success in this area is a pre-requisite for large-scale harvesting of opinions from qualitative survey data and sophisticated media analysis. The successful use of TM to integrate heterogeneous knowledge sources in biology is leading to the discovery of gene-disease associations [26]; applied more widely it will lead to TM ushering in and supporting new, data-driven research modes for e-Science.
e-Research and e-Science Goals
TM enables the e-researcher and e-scientist in knowledge discovery by finding implicit associations hidden in text. It facilitates search by providing intelligent ways to retrieve information. Semantic data derived from TM support new models of e-publishing. Scientific publications are evolving to allow more collaborative ways of communication via social networks. Rich metadata and citation analysis enriched with TM allow us to do personalised searching, extracting facts of relevance to the user.
Conclusion
NaCTeM's text mining tools and services offer numerous benefits to a wide range of users. These range from considerable reductions in time and effort for finding and linking pertinent information from large scale textual resources, to customised solutions in semantic data analysis and knowledge management. Enhancing metadata is one of the important benefits of deploying text mining services. TM is being used for subject classification, creation of taxonomies, controlled vocabularies, ontology building and Semantic Web activities. As NaCTeM enters into its second phase we are aiming for improved levels of collaboration with Semantic Grid and Digital Library initiatives and contributions to bridging the gap between the library world and the e-Science world through an improved facility for constructing metadata descriptions from textual descriptions via TM.
References
- Hey, AJC & Trefethen, A., "The data deluge: an e-science perspective". In Berman F., Fox GC., Hey AJG (eds) Grid Computing: making the global infrastructure a reality. 2003, Chichester, UK: John Wiley, pp.809-824
- Termine Web Demonstrator http://www.nactem.ac.uk/software/termine/
- Acromine Web Demonstrator http://www.nactem.ac.uk/software/acromine/
- Frantzi, K., Ananiadou, S., and Mima, H., "Automatic Recognition of Multi-Word Terms: the C/NC value method". International Journal of Digital Libraries, 2000, vol. 3:2, pp. 115-130.
- Okazaki, N. and Ananiadou, S., "Building an Abbreviation Dictionary using a Term Recognition Approach", Bioinformatics 2006 22(24):3089-3095.
- Spasi?, I., et al. "Facilitating the development of controlled vocabularies for metabolomics with text mining", 2007, in ISMB/ECCB, Bio-Ontologies SIG Workshop, Vienna, Austria, pp. 103-106
- Cheshire3-Termine Demonstration using Medline Abstracts http://www.nactem.ac.uk/software/ctermine/
- Cheshire3 Information Framework http://www.cheshire3.org/
- MEDIE http://www-tsujii.is.s.u-tokyo.ac.jp/medie/
- Miyao, Y., et al. "Semantic Retrieval for the Accurate Identification of Relational Concepts in Massive Textbases", 2006, In Proc. COLING-ACL, pp.1017-1024.
- Info-PubMed https://www-tsujii.is.s.u-tokyo.ac.jp/info-pubmed/
- ASSERT http://www.nactem.ac.uk/assert/
- Ananiadou, S., Procter, R., Rea, B., Sasaki, Y., and Thomas, J., "Supporting Systematic Reviews using Text Mining", 2007, 3rd International Conference on e-Social Science, Ann Arbor.
- Korn, N., Oppenheim, C. and Duncan, C., "IPR and Licensing issues in Derived Data", Report submitted to the JISC, April 2007
- Doyle H, Gass A, Kennison, R. "Open Access and Scientific Societies" PLoS Biology, 2004, Vol. 2, No. 5, e156: doi:10.1371/journal.pbio.002015
http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0020156 - Rowlands, I, Nicholas D., "New journal publishing models: An international survey of senior researchers". 2005 http://www.slais.ucl.ac.uk/papers/dni-20050925.pdf
- Piao, S., Ananiadou S. and McNaught, J., Integrating Annotation Tools into UIMA for Interoperability, 2007, Sixth UK e-Science All Hands Meeting (AHM2007)
- J. D. Kim, T. Ohta, Y. Tateishi, and J. Tsujii, "GENIA corpus - a semantically annotated corpus for biotext mining", Bioinformatics,19(suppl. 1):i180-i182 2003.
- e-Research Community: About the Enabling Uptake of e-Infrastructure Services Project http://www.e-researchcommunity.org/projects/e-uptake/
- Tekiner, F. and Ananiadou, S. Towards Text Mining Terabytes of Text Documents, 2007 Microsoft e-Science Workshop https://www.mses07.net/Main.aspx
- Hahn, U., Buyko, E., Tomanek, K., Piao, S., McNaught, J., Tsuruoka, Y. and Ananiadou, S. "An Annotation Type System for a Data-Driven NLP Pipeline." 2007, The Linguistic Annotation Workshop (LAW) of ACL 2007 http://www.ling.uni-potsdam.de/acl-lab/LAW-07.html
- Lally, A., and Ferrucci, D., "Building an Example Application with the Unstructured Information Management Architecture", 2004, IBM Systems Journal 43, No. 3, 455-475.
- Kano, Y., Nguyen, N., Saetre, R., Yoshida, K., Miyao, Y., Tsuruoka, Y., Matsubayashi, Y., Ananiadou, S. and Tsujii, J. (2008) Filling the gaps between tools and users: a tool comparator, using protein-protein interaction as an example, in PSB 2008, Hawaii http://psb.stanford.edu/
- Taverna Project Web site http://taverna.sourceforge.net/
- REFINE Project: Representing Evidence For Interacting Network Elements http://dbkgroup.org/refine/
- Chun, H., Tsuruoka, Y., Kim, J.D, Shiba, R., Nagata, N., Hishiki, T. and Tsujii, J. "Automatic Recognition of Topic-Classified Relations between Prostate Cancer and Genes using MEDLINE Abstracts", 2006, BMC-Bioinformatics. 7 (Suppl 3).
